Shanghai light source user research found that C / D RNA protein complex catalyzes the structural mechanism of RNA ribose methylation On January 27, the Ye Keqiang laboratory of the Beijing Institute of Life Sciences published a paper titled Structural basis for site-specific ribose methylation by box C / D RNA protein complexes in Nature. This paper studies the structural mechanism of C / D RNA protein complex catalyzing RNA ribose methylation, which is another important achievement achieved by using Shanghai Light Source Biomacromolecule Crystalline Station. Structure diagram of C / D RNA protein complex C / D RNA is a type of ancient non-coding RNA that is ubiquitous in eukaryotes and archaea. They mainly mediate the modification of ribose methylation on a large number of specific sites of ribosomal RNA and spliceosome RNA. Assembly of ribosomes of nuclear organisms. In archaea, C / D RNA forms a complex with the methyltransferase fibrillarin, RNA binding protein L7Ae and the backbone protein Nop5. The C / D RNA can complement the base sequences on both sides of the modification site to achieve specific selection of the substrate. Although there have been many studies on the structure of this compound, the two basic problems remain unsolved. First of all, how does C / D RNA assemble with protein to form a complex? The classic model thinks that one C / D RNA and two sets of proteins combine to form a so-called "monomer" structure, but recent research believes that two C / D RNA Combined with four sets of proteins to form a "cross-duplex" structure. The second question is how does C / D RNA guide methyltransferases to select specific modification sites? Using crystal X-ray diffraction data collected at the Shanghai Light Source Biomacromolecule Crystalline Station (BL17U), Ye Keqiang's laboratory analyzed a 3.15 angstrom crystal structure of a complete C / D RNA protein complex loaded with substrate. This structure shows for the first time the overall assembly of this complex molecular machine and provides direct evidence for the "monomer" model. This structure also clearly shows how the substrate RNA binds and how the catalytic subunit selects specific modification sites. The authors also found that in order to form the active state required for catalysis, the loading of the substrate induced extensive changes in the internal structure of the complex. Ye Keqiang's laboratory has been studying the structure of C / D composites for many years. Before the Shanghai light source was operated, they had collected data from synchrotron radiation sources abroad, but they have always faced a long period of application for experiments abroad, poor mobility and a long road Wait for many inconveniences. The high-brightness and stable X-rays provided by the Shanghai Light Source Biomacromolecule Crystal Beam Line Station provide great convenience and guarantee for domestic structural biologists to work in the fiercely competitive research field. They used BL17U to collect data from the beginning of the operation of Shanghai Light Source in May 2009. Due to the weak diffraction ability of the C / D composite crystal, the diffraction limit can only be reached under high-intensity synchrotron radiation X-rays. Within a year or so, they obtained a variety of crystals. With Shanghai Light Source, they can check the quality of various crystals in time and determine the next optimization direction. In May 2010, they obtained a new type of substrate-bonded composite crystal. The experiment was arranged in time through the coordination of Shanghai light source staff, and the diffraction data with a resolution of 3.15 Angstroms was obtained, so that the structure can be successfully resolved. Ye Keqiang's laboratory also studied the structure of another class of H / ACA RNA protein complex that catalyzes the formation of pseudouracil. With the analysis of the structure of the C / D RNA protein complex, the working principle of two types of complexes involved in RNA modification in the organism has been understood at the atomic level.
Kids` healthy growing-up is always parents`
most caring thing. So, we think and hope like all parents do, we want to offer
most safe and environmentally friendly kid`s furniture to all kids.
One full set of kid`s
furniture usually consists of Kids Beds, kids Night Stands, kids dressers, kids
desks, Kids Wardrobes.
To ensure kid`s
health, we are so strict with ourselves and only use the best base materials. In
order to avoid hurting the kids, we try to use less square corners as possible
in designs.
For other wooden
furniture, we offer as many as 15 wooden colors and
different sizes so that clients can fit their room well. Also, we offer size customizing
service if the quantity meeting our MOQ. Clients` safety and health is always
our priority, thus, we only use E0 standard raw materials. Besides the top
quality raw materials, we also use top-quality hardware for more durability and
stability, such as Blum, Hettich, Hafele, DTC.
Kids Beds Childrens Bunk Beds,Childrens Beds,Kids Beds,Bunk Beds Hangzhou Taihua Home Furnishing Technology Co.,Ltd , https://www.taihuafurniture.com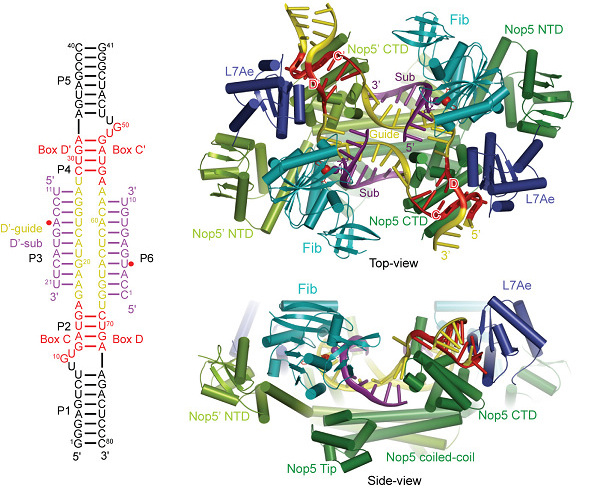
Structure mechanism of C / D RNA complex catalyzing RNA ribose methylation